What is Co-immunoprecipitation?
Protein-protein interactions play a pivotal role in cell biology and molecular biology. Understanding how proteins interact with each other to form complexes and how these interactions regulate cellular functions is crucial for unraveling biological processes. In this context, Co-immunoprecipitation (Co-IP) has emerged as a valuable biochemical technique. It allows researchers to selectively enrich proteins of interest and their interacting partners, shedding light on the critical protein-protein interactions within the cell.
Co-IP holds immense significance in molecular research due to its ability to:
Reveal Functional Insights: It helps elucidate the functional associations between proteins and unveils the dynamics of protein complexes.
Aid Drug Discovery: Co-IP plays a vital role in identifying potential drug targets and assessing the impact of drugs on specific protein interactions.
Advance Cancer Research: Dysregulated protein interactions often underlie cancer, making Co-IP a valuable tool in identifying oncogenic proteins and therapeutic targets.
Decipher Cell Signaling: Co-IP enables the capture of transient protein interactions, contributing to our understanding of intricate cell signaling pathways.
Fuel Functional Proteomics: Co-IP facilitates large-scale studies of protein functions and interactions, driving the field of functional proteomics forward.
Principles of Co-IP
Co-IP relies on several fundamental principles:
Antibody Specificity: The cornerstone of Co-IP is the use of highly specific antibodies. These antibodies are chosen based on their ability to selectively recognize and bind the target protein within a complex protein mixture.
Immobilization of Antibodies: Antibodies are immobilized onto a solid support matrix, such as Protein A/G agarose beads or magnetic beads. This immobilization maintains antibody integrity while facilitating subsequent separation steps.
Protein Binding: The biological sample, containing the target protein and potential interactors, is combined with the immobilized antibody. The antibody selectively binds to the target protein, forming an antibody-protein complex. The specificity of this binding is pivotal.
Precipitation: Following the binding step, the antibody-protein complex is separated from unbound proteins. Precipitation is typically achieved through centrifugation or magnetic separation.
Antibodies and Target Proteins for Co-immunoprecipitation
The success of Co-IP fundamentally relies on two pivotal aspects: the judicious selection of antibodies and a clear understanding of the target protein:
Antibody Selection: The choice of antibody is a critical determinant of Co-IP success. Researchers must exercise great care in selecting an antibody with a high degree of specificity and affinity for the target protein. This antibody acts as the linchpin of the Co-IP experiment, ensuring the selective capture of the target protein complex. Researchers can choose from a plethora of commercially available antibodies or opt for custom-made antibodies, weighing factors such as specificity, cross-reactivity, and accessibility.
Target Protein Identification: The core objective of Co-IP is to unravel protein-protein interactions involving a specific target protein. Therefore, a comprehensive understanding of the target protein's biological role and its anticipated binding partners is indispensable. This knowledge guides the selection of appropriate antibodies and provides valuable context for interpreting Co-IP results.
Experimental Design for Co-IP
Define Your Objective:
Clearly state the purpose of your Co-IP experiment. Are you interested in identifying binding partners of a specific protein, characterizing a protein complex, or studying a particular interaction under specific conditions?
Select Target and Control Samples:
Determine which biological samples you will use for the experiment. You'll typically need both a sample containing the target protein of interest and a control sample without the target protein to assess specificity.
Antibody Selection:
Choose an antibody with high specificity and affinity for your target protein. Verify its suitability for Co-IP through literature review or pilot experiments.
Sample Preparation:
Optimize sample preparation procedures to maintain the integrity of protein complexes. Use protease and phosphatase inhibitors to prevent degradation.
Validation Controls:
Include positive and negative controls in your experiment. Positive controls should demonstrate known interactions, while negative controls should confirm the specificity of your Co-IP.
Co-IP Procedure
Antibody Coupling:
Immobilize the selected antibody onto a solid support, such as Protein A/G agarose beads or magnetic beads, following the manufacturer's guidelines.
Validate the efficiency of antibody coupling using control samples.
Pre-clearing:
Incubate your prepared sample with a control resin to remove non-specifically binding proteins. This step reduces background noise.
Incubation with Antibody Beads:
Mix the pre-cleared sample with the immobilized antibody beads and incubate under gentle agitation. Allow sufficient time for specific antibody-target protein binding to occur (usually a few hours or overnight).
Washing:
Carefully wash the antibody-protein-bead complex to remove unbound proteins and contaminants. Use stringent wash buffers to enhance specificity.
Elution:
Elute the captured protein complexes from the antibody beads using an appropriate elution buffer. This step releases the bound proteins for downstream analysis.
Analysis:
Analyze the eluted samples using various techniques, such as SDS-PAGE, Western blotting, or mass spectrometry, depending on your research objectives.
Confirm the presence of the target protein and its interacting partners.
Data Interpretation:
Interpret your results based on the presence or absence of specific protein-protein interactions. Compare results with controls to assess specificity.
Validation:
Validate your findings through independent experiments, alternative techniques, or functional assays, if applicable.

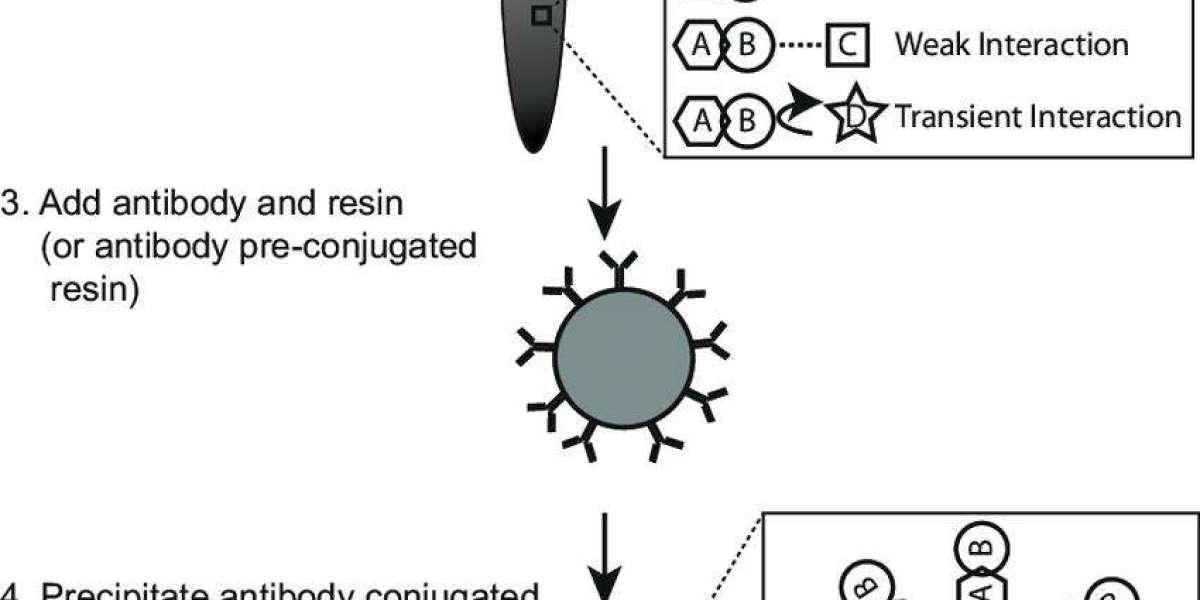
Schematic procedure of coimmunoprecipitation (co-IP) assay (Tang et al., 2018).
Quality Control:
Consistently monitor and document all experimental conditions, including incubation times, temperatures, and buffer compositions.
Ensure the specificity of your Co-IP by comparing results with appropriate controls.
Use high-quality antibodies and reagents to minimize non-specific binding and false positives.
Application of Co-Immunoprecipitation (Co-IP) in Scientific Research
Protein-Protein Interaction Studies:
Co-immunoprecipitation (Co-IP) is a fundamental technique used to identify and characterize protein-protein interactions. It helps unravel the composition of protein complexes and their roles in cellular processes. In cancer research, Co-IP revealed the interaction between the tumor suppressor p53 and the E3 ubiquitin ligase MDM2, shedding light on p53's regulation.
Cell Signaling Pathways:
Co-IP is a crucial tool for dissecting intricate cell signaling pathways. It assists in mapping protein interactions and pinpointing key components in signal transduction cascades. Studies using Co-IP have unveiled interactions within the Wnt signaling pathway, like those involving β-catenin and GSK-3β, offering insights into cell proliferation and differentiation regulation.
Epigenetics and Chromatin Remodeling:
Co-IP is employed to delve into epigenetic modifications and chromatin-associated proteins, revealing their roles in gene regulation and epigenetic control. Investigations using Co-IP have unveiled interactions between histone modification enzymes and specific histone marks, providing understanding of gene expression regulation.
Disease Mechanisms and Biomarker Discovery:
Co-IP is vital for unraveling the molecular mechanisms behind diseases and discovering potential biomarkers. For instance, in Alzheimer's disease research, Co-IP has been pivotal in identifying protein interactions associated with the disease, such as the aggregation of tau and amyloid-beta proteins.
Drug Target Validation:
Co-IP plays a pivotal role in validating drug targets by confirming physical interactions between drug candidates and their intended protein targets. It is utilized to validate the binding of small molecules to specific kinase proteins in drug discovery efforts.
Functional Proteomics:
Co-IP facilitates the study of protein interactions within cellular organelles, providing insights into organelle-specific functions and processes. It has been instrumental in elucidating protein interactions within mitochondria, revealing dynamics of mitochondrial protein complexes involved in energy production.
Virus-Host Interactions:
Co-IP is a valuable tool for studying interactions between viral proteins and host cellular proteins. It has identified host factors interacting with viral proteins during HIV infection, contributing to our understanding of potential antiviral targets.
RNA-Protein Interactions:
Co-IP is adapted to investigate interactions between RNA molecules and RNA-binding proteins (RBPs). It has unveiled RBPs binding to specific non-coding RNAs, impacting gene regulation in the field of RNA biology.
Stem Cell Biology and Differentiation:
Co-IP is used to study protein interactions involved in stem cell maintenance, differentiation, and reprogramming. It has identified protein complexes associated with pluripotency factors, offering insights into stem cell fate determination.
Cell Cycle Regulation:
Co-IP is applied to investigate interactions between cell cycle regulatory proteins. It has elucidated interactions between cyclins and cyclin-dependent kinases (CDKs), key components governing cell cycle control.
Reference
- Tang, Zhenyuan, and Yoshinori Takahashi. "Analysis of protein–protein interaction by co-IP in human cells." Two-Hybrid Systems: Methods and Protocols (2018): 289-296.